Welcom to muti-species TR and IG Database !
Introduction
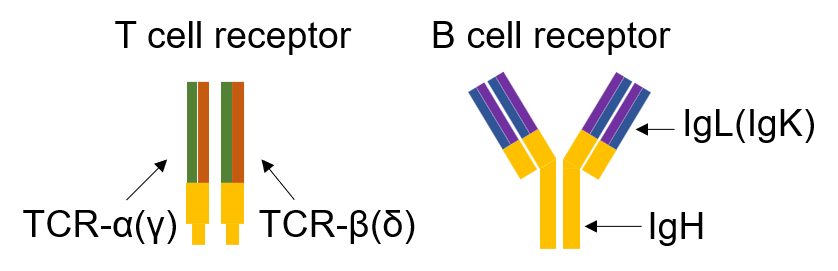
T cell receptor (TCR) is composed of two different protein chains. The vast majority of T cells express TCRs composed of α (alpha) and β (beta) chains while a small subset expresses TCR composed of γ (gamma) and δ (delta) chains. The sequence encoded by the V(D)J junction is called complementarity determining region 3 or CDR3. This sequence has the highest variability in both alpha and beta chains and determines the ability of a T cell to recognize an antigen peptide presented by the MHC molecule.
Each B cell expresses a single B cell receptor (BCR), and the diverse range of BCRs expressed by the total B cell population of an individual is termed the 'BCR repertoire'. Many B cells undergo 'isotype switching', in which stepwise DNA deletion and recombination from immunoglobulin M (IgM) generates downstream isotypes (IgG1, IgG2, IgG3 and IgG4, IgA1 and IgA2, IgD and IgE) that confer distinct functional characteristics and roles in disease.
News
2023.12.12: TRIgdb version v1.0 has published.
2024.05.30: TRIgdb version v2.0 has published. We have added functionality for users to upload their own data for analysis in the Clonal Analysis page.
2025.06.10: TRIgdb version v3.0 has published. We have implemented a DeepSeek-powered search feature in Chat page.
Overview
TRIgdb contains 497 samples from 12 species. For each sample, the database provides the immune repertoire information, including TCRA, TCRB, TCRD, and TCRG, as well as antibodies such as IgG, IgH, IgM, IgA, and others. The recovery of the immune repertoire is derived from bulk RNA-seq using the TRUST software. Additionally, the website also offers CDR3 sequence alignment functionality, as well as analysis of immune repertoire diversity and richness for each sample or species.
Function
1. Download TR and IG tables and meta information for 12 species.
2. Providing CDR3 sequence alignment functionality.
3. Analysis of immune repertoire for 12 species.
4. Upload your data for immune repertoire analysis.
5. Providing DeepSeek-powered search functionality across 3.55 × 105 TCR and 2.90 × 106 BCR sequences.
TR and IG tab
Data Upload for Analysis
File Upload Format Requirements:
Choose Files(.tsv):
File format: Files must have the .tsv extension.
File content: Files must contain at least the following columns: clone count (Count), amino acid sequence (CDR3Aa), nucleotide sequence (CDR3nt), V, J, C. If conducting group comparisons, it is recommended to have a minimum of 2 samples per group.
Choose Files(metadata.xlsx)
File format: Metadata table must have the .xlsx extension
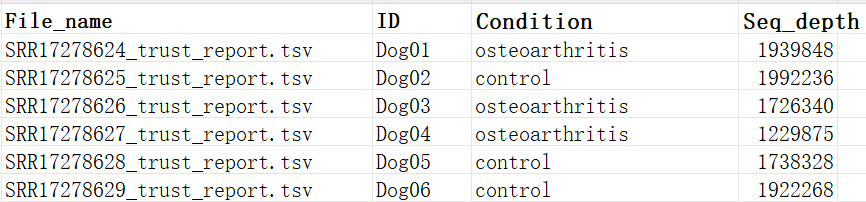
File content: Metadata table must contain at least the following columns: file name (File_name), sample ID (ID). It can also include grouping information (Condition) and sample sequencing depth (Seq_depth).
Remove batch effects:
Raw_count: Use the count values from the .tsv file for further analysis.
Sequencing_Depth: Ensure that the Metadata table includes a column for sequencing depth (Seq_depth) for each sample.
TRIG_Count: Use the number of immune repertoire libraries to remove batch effects.
Group or not:
If you need to perform group comparisons, please ensure that the Metadata table includes a column for grouping information (Condition) to enable group comparisons.
Analysis content:
Clonotype Analysis
Gene Uasge Analysis
Diversity Analysis
Please prepare the files for upload according to the above requirements.
Here is an example: a .tsv file and a metadata file
.tsv file:

metadata.xlsx file:
Download
TRIgdb includes 497 samples from 12 species.
We counted the number of TR and IG genes for each sample and collected the meta-information for each sample.
The TR and IG tables contain 16 columns, which are:
File_name
Species
ID
Status
Number_of_Seq
Number_of_TRA
Number_of_TRB
Number_of_TRD
Number_of_TRG
Number_of_IGHA
Number_of_IGHG
Number_of_IGHM
Number_of_IGHD
Number_of_IGHY
Number_of_IGL
Number_of_IGK
Download TR and IG TabMetadata table contain 10 columns, which are:
ID
Condition:grouping information
Tissue
Species name
Sex
Age
Breed or Chinese name
Breeding method
Sampling Time
Datasets
Download metadata Tab